Tissue specificity in mucosal-microbiome interactions
Research Theme 3
About
It is now clear that tissue-resident immune cells exhibit specialised responses according to the tissue environment.
For example, intestinal macrophages display some common but some distinct features compared to macrophages in other tissues.1Takata K et al., Induced-Pluripotent-Stem-Cell-Derived Primitive Macrophages Provide a Platform for Modeling Tissue-Resident Macrophage Differentiation and Function. Immunity, 2017 47:183-198 e6
For example, E. coli strains that exist commensally in the gastrointestinal tract may be opportunistic pathogens in the female reproductive tract and pathogens within the urinary tract.
3.1
Tissue Specific Macrophages and Epithelial Cells
A transcriptome atlas of human tissue resident macrophages in the Stemformatics platform has previously been assembled
that allows users to navigate the expression of key pathogen recognition receptors, and highlight differences in inflammatory or metabolic pathways.
This atlas will be further developed using RNAseq to compare tissue specific responses of Mφ/DC to the human microbiome. Using tissue specific human-derived microbiomes will provide new knowledge on the innate responses and the function of the microbiome at different mucosal sites, which is essential for the development of tissue specific microbiome medicines or probiotics.
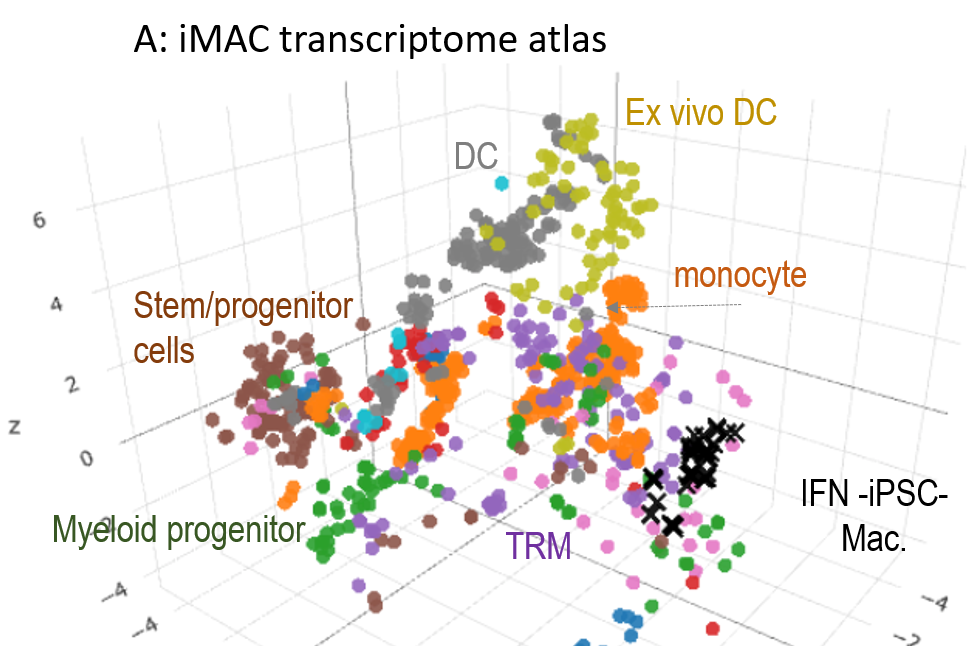
The integration of transcriptome data describing the phenotypes of human tissue resident macrophages, monocytes and iPSC-derived macrophages ± IFN. The atlas allows identification of key genes or gene networks that discriminate between the type of Mφ and its response to pathogen B.
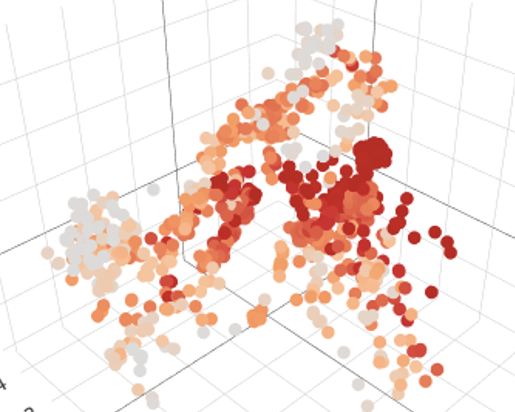
An example of restricted expression of the pattern recognition receptor Clec4ein monocytes and iPSC-derived Mφ.
3.2
Measuring the tissue specific commensal responses to native and foreign host cells
We will create a transcriptional atlas to explore host-microbiome interactions, providing a visual interaction point for the underlying multi-omics data and facilitating data exploration by the scientific community.
Outcome
The integration of our transcriptome data, with maps of human tissue resident innate immune cell phenotypes, provides an important resource for identifying and targeting mucosal cell populations. Machine learning to identify key innate immune cell behaviours will provide a nuanced understanding of the interaction between site-specific mucosal cells and microbiome communities. The availability of the data in the Stemformatics platform increases usability and exploration by the broader scientific community.
Our Research Themes
Innate immune sensing of the human intestinal microbiome
Theme 1
Innate immune shaping of the intestinal microbiome
Theme 2
Tissue specificity in mucosal-microbiome interactions
Theme 3
Disruption of mucosal-microbiome interactions
Theme 4
Get the latest updates
Subscribe to our mailing list to keep up to date with the latest news from Synnate
